Software
The list below highlights my most prominent software contributions to Artificial Life in general. Recent libraries/frameworks are coded in Python for maximal inter-operability while older ones are expected to be either ported or binded in the near future.
AMaze
 A lightweight maze navigation task generator for sighted AI agents.
This library is meant to serve as a fast-prototyping platform to test various algorithms while being:
a) computationally cheap; b) unbounded in complexity; and c) intuitively understandable by a human.
The library works with either fully discrete perception and action spaces; fully continuous; or with continuous perceptions and discrete actions.
A lightweight maze navigation task generator for sighted AI agents.
This library is meant to serve as a fast-prototyping platform to test various algorithms while being:
a) computationally cheap; b) unbounded in complexity; and c) intuitively understandable by a human.
The library works with either fully discrete perception and action spaces; fully continuous; or with continuous perceptions and discrete actions.
Multiple investigations into its properties are detailed on its project page, join in!
Available as a pure-python library on pypi.
More information on read the docs.
ABrain
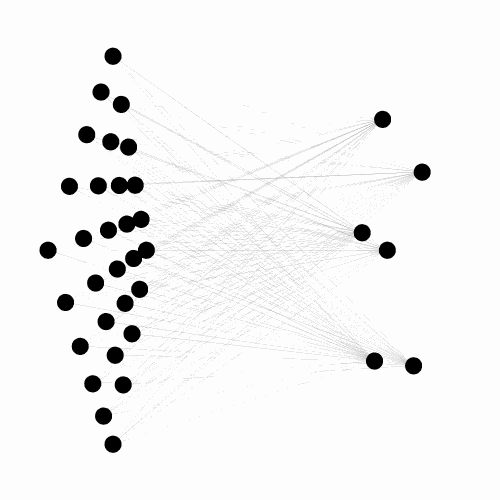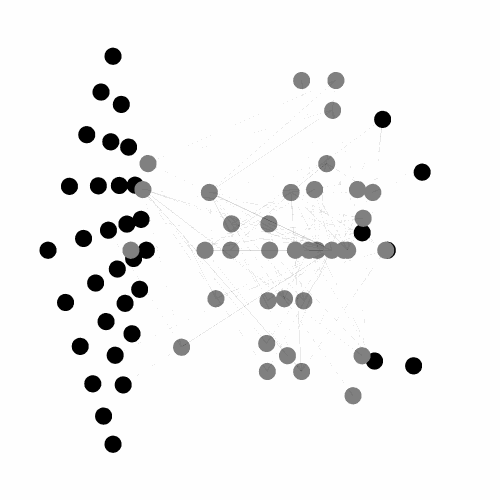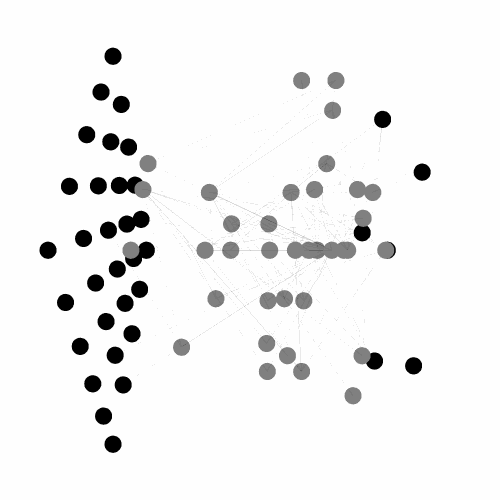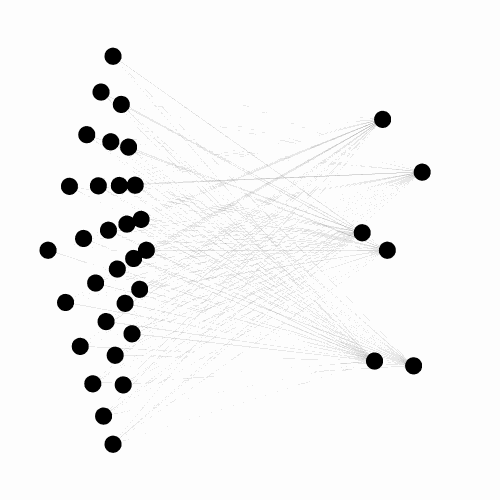 A C++ library, with python binding, implementing the Evolvable Substrate Hyper NeuroEvolution through Augmenting Topologies (ES-HyperNEAT) algorithms.
Works, by default, on three dimensional substrates and can handle large input/output spaces.
Also provides numerous tools for visualizing both the 3D ANN (dynamically or not) and the underlying CPPN. Currently under active development, see the related project for more details.
A C++ library, with python binding, implementing the Evolvable Substrate Hyper NeuroEvolution through Augmenting Topologies (ES-HyperNEAT) algorithms.
Works, by default, on three dimensional substrates and can handle large input/output spaces.
Also provides numerous tools for visualizing both the 3D ANN (dynamically or not) and the underlying CPPN. Currently under active development, see the related project for more details.
Available on pypi.
More information on read the docs.
Splinoids
 A C++ framework for modeling 2D robots with evolvable morphological components based on cubic splines, hence the name.
The robots are endowed with vision, touch and hearing; can locomote via two wheels and speak on multiple frequencies.
When morphologically appropriate, they also have control over each articulation allowing them to use their arms offensively or defensively.
A C++ framework for modeling 2D robots with evolvable morphological components based on cubic splines, hence the name.
The robots are endowed with vision, touch and hearing; can locomote via two wheels and speak on multiple frequencies.
When morphologically appropriate, they also have control over each articulation allowing them to use their arms offensively or defensively.
Available on github.
APOGeT
 A C++ tool for producing phylogenetic trees from genealogical data.
The Automated Phylogeny Over Geological Timelines uses a (evolvable) genetic distance metric to separate species.
The underlying algorithms rely on a methaphorical membrane that is extended through genetic divergence until a child species is created from its parent.
The methology has been used for both asexual and sexual reproductions and can handle most corner cases such as hybridation from distant species.
A C++ tool for producing phylogenetic trees from genealogical data.
The Automated Phylogeny Over Geological Timelines uses a (evolvable) genetic distance metric to separate species.
The underlying algorithms rely on a methaphorical membrane that is extended through genetic divergence until a child species is created from its parent.
The methology has been used for both asexual and sexual reproductions and can handle most corner cases such as hybridation from distant species.
Available on github.
Citation